Any scientist with access to the news is excited by the possibilities implicit in the amazing advances in genetics research in the recent decade, even up here in Georgia’s Appalachia. And each scientist puts their own twist on what might be up ahead. In Psychiatry, genetic markers have long been suspected to play a role in Schizophrenia , Manic Depressive Illness, Attention Deficit Disorder, etc. The possibility of finding those connections preoccupies many. But there are a lot of neuroscientists who think that there are genetic explanations that explain the variable response to medications seen with most psychopharmacologic agents. So the race is on to find those connections. I’ve previously looked at this study, but I know more now, so I want to revisit it. The full text of this article is available online here:
Prediction of Antidepressant Response to Milnacipran by Norepinephrine Transporter Gene Polymorphisms
by Keizo Yoshida, Hitoshi Takahashi, Hisashi Higuchi, Mitsuhiro Kamata, Ken-ichi Ito, Kazuhiro Sato, Shingo Naito, Tetsuo Shimizu, Kunihiko Itoh, Kazuyuki Inoue, Toshio Suzuki, Charles B. Nemeroff
American Journal of Psychiatry 2004; 161:1575–1580
Abstract:Objective: With a multitude of antidepressants available, predictors of response to different classes of antidepressants are of considerable interest. The purpose of the present study was to determine whether norepinephrine transporter gene (NET) and serotonin transporter gene (5-HTT) polymorphisms are associated with the antidepressant response to milnacipran, a dual serotonin/norepinephrine reuptake inhibitor.Method: Ninety-six Japanese patients with major depressive disorder were treated with milnacipran, 50–100 mg/day, for 6 weeks. Severity of depression was assessed with the Montgomery-Åsberg Depression Rating Scale. Assessments were carried out at baseline and at 1, 2, 4, and 6 weeks of treatment. The method of polymerase chain reaction was used to determine allelic variants.Results: Eighty patients completed the study. The presence of the T allele of the NET T-182C polymorphism was associated with a superior antidepressant response, whereas the A/A genotype of the NET G1287A polymorphism was associated with a slower onset of therapeutic response. In contrast, no influence of 5-HTT polymorphisms on the antidepressant response to milnacipran was detected.Conclusions: The results suggest that NET but not 5-HTT polymorphisms in part determine the antidepressant response to milnacipran.
96 Japanese patients with Major Depressive Disorder were treated with an SRNI [malnicipran]. Since we are sure to be bombarded with biomarkers studies, I thought I’d run through a few quick steps for vetting them as they come along.
Who paid for the study?
"Supported in part by grants from the Mitsubishi Pharma Research Foundation and Asahi Kasei Pharma."
Malnicipram is manufactured by Asahi Kasei.
Independent of the genetics, is the study viable?
- The patients were treated for six weeks but there are no placebo controls. We know from the plethora of clinical trials with antidepressants that the placebo effect can be quite large so it’s hard/impossible to evaluate how much of the improvement in scores was drug effect. Likewise they "digitized" their results into responders and nonresponders based on a 50% reduction in scores rather than compare the raw data itself.
Is the genetic material presented or only described?
Good for them! They showed us their data.



On the left T/T, T/C, and C/C for NET T-182C
In the Center G/G, G/A, and A/A for NET G1287A
On the right S/S, S/L, and L/L for 5-HTTLPR
I don’t see much there. The only significance they saw was between G/A and A/A [p=0.04]
Were there any ‘magic tricks’ with the data?
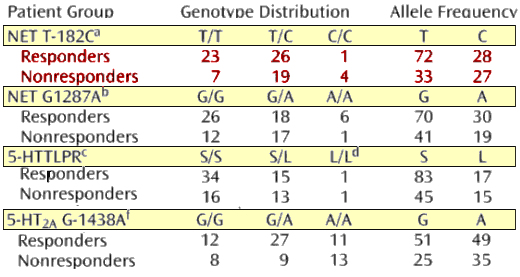
You bet! They turned the non-significant difference in NET T-182C [graph on the left] into responders and non-responders based on a 50% drop in MADRS score which made it look a lot different! They then found the data [in red] significant [p=0.03].
Did they correct for the number of alleles they tested?
- No! They tested five alleles, so since their series level of significance was p<0.05, with n=5 they would need p<0.01 for significance on any given allele [0.05/5 = 0.01]. All they got was p=0.03. [see personalized medicine: the shoals of fuzzy math…]
So, this is an company sponsored study on its own drug to determine if various alleles of the noradrenaline and serotonin transporter genes predicted an antidepressant response. Since there was no placebo group, we’re not even sure if there actually was any antidepressant response, but we can certainly see in the graphs and their statistics that there were no apparent robust differences among alleles. And even though they achieved "significance" by jury-rigging the data by digitizing it, the reported significance disappeared when the data was corrected for multiple tests. This study should not have been published, particularly in a prestigious journal like the American Journal of Psychiatry. I’m not going through this study to scold anyone. It was published in 2004, light years ago in genome time. Back then, who knew? But, now it’s 2011 and people are up to speed about genetic studies:
- We do know that the data needs to be corrected for the number of alleles tested.
- We do know to look at the quality of the study independent of the genetics.
- We do know to look at magic conversions that ‘create significance.’
- We do know to be careful about industry sponsored trials in general.
- We do know to look at the authors’ conflicts of interest [unaddressed in this paper!].
- We do know to look for ‘editorial support’ [ghost-writing].
-
We do know how much people want to move into these lucrative ‘personalized’ markets. We should expect a lot of fudging in such studies.
Of course we should be vigilant with all we now know when we read our literature about genetic studies. But the Journal editors know all of these things too. It’s incumbent on them to thoroughly vet the literature they publish. If they don’t, it’s incumbent on us to raise holy hell about it. In my opinion, it’s part of the Hippocratic Oath to keep our literature scientifically honest, particularly in an area like this one that’s so rife for mangled science. This study, in my opinion, is a preconceived conclusion grasping for an argument to support it. Since none emerged from the study, they moved the study to meet the conclusion. This statement is unsupported by anything rational, "Conclusions: The results suggest that NET but not 5-HTT polymorphisms in part determine the antidepressant response to milnacipran." We slept through a lot of similar sheenanigans back in the last twenty plus years to our great shame, mine included. We can’t let that happen again…
It gets even worse. Notice that Charles Nemeroff is an author with these Japanese investigators. He even manipulated the credit line to say the study is from Emory, ahead of the Japanese identifiers. Nemeroff was outed in 2003 for failing to disclose his financial interest in milnacipran, among other omissions, in a Nature Neuroscience review article. The New York Times broke the story and Nature Publishing Group finally revised their policies. That was the beginning of Nemeroff’s fall from grace. So here we have this article in 2004 that carries no disclosure about Dr. Nemeroff’s interest in the drug.
To the best of my knowledge Dr. Yoshida in Japan is a reputable investigator. Who put in the fix for Dr. Nemeroff to insinuate himself into this Japanese project? Was it Dr. Nemeroff’s job to spin the data at the behest of the corporation?
I agree that the “digitization” is pure hocus-pocus. Patients whose symptoms improve by 51% generally don’t feel all that better than those who improve by 49%.
Proof once again that, if you torture your statistics long enough, they will confess to anything.