Overview of the Genetics of Major Depressive Disorder
Falk W. Lohoff
Current Psychiatry Reports. 2010 12[6]: 539–546.
[full text on-line]
Abstract: Major depressive disorder [MDD] is a common psychiatric illness with high levels of morbidity and mortality. Despite intensive research during the past several decades, the neurobiological basis and pathophysiology of depressive disorders remain unknown. Genetic factors play important roles in the development of MDD, as indicated by family, twin, and adoption studies, and may reveal important information about disease mechanisms. This article describes recent developments in the field of psychiatric genetics, with a focus on MDD. Early twin studies, linkage studies, and association studies are discussed. Recent findings from genome-wide association studies are reviewed and future directions discussed. Despite all efforts, thus far, no single genetic variation has been identified to increase the risk of depression substantially. Genetic variants are expected to have only small effects on overall disease risk, and multiple genetic factors in conjunction with environmental factors are likely necessary for the development of MDD. Future large-scale studies are needed to dissect this complex phenotype and to identify pathways involved in the etiology of MDD.
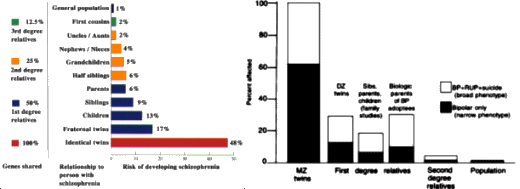
Major depressive disorder [MDD] is a common psychiatric illness with high levels of morbidity and mortality. It is estimated that 10% to 15% of the general population will experience clinical depression during their lifetime [1], and 5% of men and 9% of women will experience a depressive disorder in a given year, according to the World Health Organization [2]. Genetic factors play important roles in the development of MDD, as indicated by family, twin, and adoption studies. Twin studies suggest a heritability of 40% to 50%, and family studies indicate a twofold to threefold increase in lifetime risk of developing MDD among first-degree relatives. This degree of familial aggregation, coupled with the high heritability from twin studies, generated optimism that molecular genetic techniques would reveal genes of substantial influence on MDD risk. Unfortunately, gene localization and identification has been a slow, labor-intensive process. Genetic investigators have encountered similar frustrations with other common complex traits [eg, asthma, hypertension, and diabetes mellitus].
The major impediments to mood disorder gene localization and identification are as follows: [1] no single gene is necessary and sufficient for MDD; [2] each susceptibility gene contributes a small fraction of the total genetic risk; and [3] complex genetic heterogeneity, meaning that multiple partially overlapping sets of susceptibility genes [which interact with the environment] can predispose individuals to similar syndromes that are indistinguishable on clinical grounds.The field of psychiatric genetics in general has been disappointing given that the initial hope to find common gene variants of large effect in the pathogenesis of mental illnesses has been unsuccessful. In most psychiatric illnesses, the phenotype seems too complex, with the patient cohorts too small, and no findings have been consistently replicated. This is also the case for MDDs. In addition, the phenotypic effects of genetic variants identified to date are weak, with ORs of 1.0 to 1.2. The picture is further complicated when comparing the magnitude of the impact of gene variation on disease susceptibility with the impact of lifestyle and environmental factors, which is likely to be large. Despite these obstacles, the field of psychiatric genetics is rapidly growing, and several new technological advances [eg, whole-genome sequencing] will be soon available for large-scale studies. It is important to remember that genetic information will only provide additional information on one aspect of the complex and personal history of psychiatric patients. It is the sum of inside and outside factors that contributes and influences mental pathology and well-being.
-
DSM-5: Throughout the months of watching the DSM-5 implode, I’ve wondered the same thing over and over. In 2002, the future leaders of the DSM-V Task Force published a book, A Research Agenda for DSM-V, that said in the introduction:The descriptive approach adopted by the DSM allowed for the development of a classification system that met the field’s need for a common language, without being mired in ideological hypotheses about the causes of psychiatric illness. Questions have been raised by many critics that the DSM’s descriptive approach may have outlived its usefulness and is in fact potentially misleading. Although there is a large body of research that indicates a neurobiological basis for most mental disorders, the DSM definitions are virtually devoid of biology. Instead, DSM-IV definitions are based on clusters of symptoms and characteristics of clinical course.
 It is our goal to translate basic and clinical neuroscience research relating brain structure, brain function, and behavior into a classification of psychiatric disorders based on etiology and pathophysiology.While it’s not my main point here, I don’t personally think that "there is a large body of research that indicates a neurobiological basis for most mental disorders." But I sure don’t question that the body psychiatric believes that and has for some time. My preoccupation has been "What made the DSM-V [DSM-5] Task Force leaders think that they could come out of the closet and declare that the DSM-5 would aim to be based on ‘etiology and pathophysiology’?" They didn’t have anything to back up that statement. I recall the party line back in 1980 was that etiology would be added when it was known. It sure wasn’t known in 2002. And they didn’t say they would classify some psychiatric disorders by adding neurobiological data, they said "a classification of psychiatric disorders based on etiology and pathophysiology" was the goal. They stepped way out of the closet. They hedged their bets by saying such a classification might be in the far distant future, but they were pretty sure that was the future. They also thought the "descriptive approach may have outlived its usefulness" e.g. they had already given up on the DSM-III, DSM-IIIR, DSM-IV solution and were headed for greater things.
It is our goal to translate basic and clinical neuroscience research relating brain structure, brain function, and behavior into a classification of psychiatric disorders based on etiology and pathophysiology.While it’s not my main point here, I don’t personally think that "there is a large body of research that indicates a neurobiological basis for most mental disorders." But I sure don’t question that the body psychiatric believes that and has for some time. My preoccupation has been "What made the DSM-V [DSM-5] Task Force leaders think that they could come out of the closet and declare that the DSM-5 would aim to be based on ‘etiology and pathophysiology’?" They didn’t have anything to back up that statement. I recall the party line back in 1980 was that etiology would be added when it was known. It sure wasn’t known in 2002. And they didn’t say they would classify some psychiatric disorders by adding neurobiological data, they said "a classification of psychiatric disorders based on etiology and pathophysiology" was the goal. They stepped way out of the closet. They hedged their bets by saying such a classification might be in the far distant future, but they were pretty sure that was the future. They also thought the "descriptive approach may have outlived its usefulness" e.g. they had already given up on the DSM-III, DSM-IIIR, DSM-IV solution and were headed for greater things.A Research Agenda for DSM-V had an army of contributors, so it must have been in production around the time the Human Genome Project was finally bearing fruit. I think that the coming of the technology of the Human Genome Project was the thing that made them think that they could get away with finally saying that psychiatry = medicine = biology, a twenty year old dream. No more equivocating. Robins and Guze, neoKraepelinian tenets, and John Feighner set free! They had the runs in families data; they were going to finally see the genetic code and find the offending genes; and they had ten years for it to happen. It was in the bag!
I doubt that in 2000, anyone had any idea how the the cracking of the human genome wasn’t going to be the end of something, but rather a new beginning. And I think this statement above – "The field of psychiatric genetics in general has been disappointing given that the initial hope to find common gene variants of large effect in the pathogenesis of mental illnesses has been unsuccessful" – is why the DSM-5 went so far off track. They thought there would be enough in the genetic findings to validate their biologizing psychiatry formally. At least that’s my guess. Bad call! They were in too big a hurry. They hadn’t planned on "The picture is further complicated when comparing the magnitude of the impact of gene variation on disease susceptibility with the impact of lifestyle and environmental factors, which is likely to be large." -
Translational Science: "Translational science is a cross disciplinary, scientific research that is motivated by the need for practical applications that help people. The term is used mostly in the health sciences and refers to real-time translation of bench science, conducted only in a lab, to bedside clinical practice or dissemination to population-based community interventions." While it sounds good, it’s a racket. Make your research sound practical, and you’ll get funded. $35 M for STAR*D is one of the reasons I don’t like it. Genetic research like this is an example:Common Genetic Variation and Antidepressant Efficacy in Major Depressive Disorder: A Meta-Analysis of Three Genome-Wide Pharmacogenetic Studies
by the GENDEP Investigators, MARS Investigators, and STAR*D Investigators
American Journal of Psychiatry. 2013 170:207–217.Objective: Indirect evidence suggests that common genetic variation contributes to individual differences in antidepressant efficacy among individuals with major depressive disorder, but previous studies may have been underpowered to detect these effects.Method: A meta-analysis was performed on data from three genome-wide pharmacogenetic studies – the Genome-Based Therapeutic Drugs for Depression [GENDEP] project, the Munich Antidepressant Response Signature [MARS] project, and the Sequenced Treatment Alternatives to Relieve Depression [STAR*D] study – which included 2,256 individuals of Northern European descent with major depressive disorder, and antidepressant treatment outcomes were prospectively collected. After imputation, 1.2 million single-nucleotide polymorphisms were tested, capturing common variation for association with symptomatic improvement and remission after up to 12 weeks of antidepressant treatment.Results: No individual association met a genome-wide threshold for statistical significance in the primary analyses. A polygenic score derived from a meta-analysis of GENDEP and MARS participants accounted for up to approximately 1.2% of the variance in outcomes in STAR*D, suggesting a weakly concordant signal distributed over many polymorphisms. An analysis restricted to 1,354 individuals treated with citalopram [STAR*D] or escitalopram [GENDEP] identified an intergenic region on chromosome 5 associated with early improvement after 2 weeks of treatment.Conclusions: Despite increased statistical power accorded by meta-analysis, the authors identified no reliable predictors of antidepressant treatment outcome, although they did identify modest, direct evidence that common genetic variation contributes to individual differences in antidepressant response.If we can’t yet identify the genetics in diseased like Schizophrenia, Manic Depressive Illness, or Major Depressive Disorder, I doubt we’re going to find the genetics in speculative hypotheses like this endless talk of personalized medicine. It’s not just this meta-analysis that I’m talking about, it’s that the NIMH has already wasted our research money on STAR*D, CO-MED, IMPACT, following this same goal of trying to find a way to get more out the antidepressants than they have to give. Now, even with this current meta-analysis coming up empty handed, we’re still funding these same investigators for EMBARC – yet another Translational Science program to predict response to antidepressant medications based on genetic profiles. In my view, we’re not there yet, but because these projects have been pitched as practical [Translational], they get funded over and over in various incarnations. EMBARC is going to come out just like STAR*D, CO-MED, and IMPACT – lost in translation.
There’s a large body of research speculating that biological or genetic markers might be found for psychiatric disorders. Good enough for government funding!
GENDEP, MARS, STAR*D, EMBARC – it’s all cargo cult science, to use Richard Feynman’s wonderful description. They go through the motions of real research, they squander millions of dollars of public research funding, and all they have to show for it is 1.2% of the variance. Such a deal! A cynic might say they are just playing the game of keeping the game going. NIMH needs new direction, soon.
The work of Jay Joseph is relevant here.
http://jayjoseph.net/yahoo_site_admin/assets/docs/2012_Joseph_Missing_Heritability_ADS_As_Published_Online.114214811.pdf
My heartfelt thanks for lucid posts and links to more, that make this old lady wide awake into the wee hours.
“While it (translational science) sounds good, it’s a racket. Make your research sound practical, and you’ll get funded.”
Years of “Early (tidlig) Intervention in Psychosis, TIPS here, aimed at shorter duration of untreated psychosis, DUP everywhere, has metamorphised into POP, the Norwegian Primary Prevention of Psychosis Project, very well funded by public means as prioritized scientific research, although the 10-year evaluation mirrors the 5-year evaluation, and something “eludes” doctors of medicine, gotten there by way of this research. When facts (patients relaps) do not support the hypothesis of treating diagnosable pre-psychosis (with drugs) to avoid psychosis, they carry on treating/medicating, avoiding the fundamental questions, the faulty thinking, the harmful drugs.
A racket, indeed, serving those with power and responsibility: Doctors, Health as Big Business, bureaucrats, researchers, parents, politicians, at the expense and the lives of vulnerable humans. Every psychiatrically diagnosed young person I’ve known had terrible histories of trauma and emotional neglect. Many are dead, others are visibly harmed by poverty and drugs. Two former patients I know are fine, free of meds, thanks to their own rejection of bipolar and schizophrenia diagnoses and solid support from parents loving enough to trust their children more than authoritarian shrinks in Norway’s publicly financed psychiatry, at the European top in use of coercive treatment/medication.