by Javier Arnedo, M.S.; Dragan M. Svrakic, M.D., Ph.D.; Coral del Val, Ph.D.; Rocío Romero-Zaliz, Ph.D.; Helena Hernández-Cuervo, M.D.; Molecular Genetics of Schizophrenia Consortium; Ayman H. Fanous, M.D.; Michele T. Pato, M.D.; Carlos N. Pato, M.D., Ph.D.; Gabriel A. de Erausquin, M.D., Ph.D.; C. Robert Cloninger, M.D., Ph.D.; and Igor Zwir, Ph.D.American Journal of Psychiatry. Published in advance on September 15, 2014.
Objective: The authors sought to demonstrate that schizophrenia is a heterogeneous group of heritable disorders caused by different genotypic networks that cause distinct clinical syndromes.Method: In a large genome-wide association study of cases with schizophrenia and controls, the authors first identified sets of interacting single-nucleotide polymorphisms [SNPs] that cluster within particular individuals [SNP sets] regardless of clinical status. Second, they examined the risk of schizophrenia for each SNP set and tested replicability in two independent samples. Third, they identified genotypic networks composed of SNP sets sharing SNPs or subjects. Fourth, they identified sets of distinct clinical features that cluster in particular cases [phenotypic sets or clinical syndromes] without regard for their genetic background. Fifth, they tested whether SNP sets were associated with distinct phenotypic sets in a replicable manner across the three studies.Results: The authors identified 42 SNP sets associated with a 70% or greater risk of schizophrenia, and confirmed 34 [81%] or more with similar high risk of schizophrenia in two independent samples. Seventeen networks of SNP sets did not share any SNP or subject. These disjoint genotypic networks were associated with distinct gene products and clinical syndromes [i.e., the schizophrenias] varying in symptoms and severity. Associations between genotypic networks and clinical syndromes were complex, showing multifinality and equifinality. The interactive networks explained the risk of schizophrenia more than the average effects of all SNPs [24%].Conclusions: Schizophrenia is a group of heritable disorders caused by a moderate number of separate genotypic networks associated with several distinct clinical syndromes.
 As much as I like to talk about Data Transparency and try to demystify some of the calculations used in the Clinical Trials, when it comes to the modern genetic studies, I’m lost at sea. They’re doing calculus and I’m still on the multiplication tables just over counting on my fingers. I know that GWAS stands for genome-wide association study, and I sort of know what a SNP [single-nucleotide polymorphism] is. Beyond that, I’m only dimly able to understand how a given study was conducted and what the authors make of their findings, but not able to speak to the quality of the work. So in this case, I’ll say what a SNP is, because these geneticists can talk of little else, and if you talk genetics, you have to throw in SNP [pronounced "snip"] every so often to qualify yourself. Remember that the genome [the sequence of nucleotides on the chromosomes] fills a pretty large cabinet and makes for some really monotonous reading.
As much as I like to talk about Data Transparency and try to demystify some of the calculations used in the Clinical Trials, when it comes to the modern genetic studies, I’m lost at sea. They’re doing calculus and I’m still on the multiplication tables just over counting on my fingers. I know that GWAS stands for genome-wide association study, and I sort of know what a SNP [single-nucleotide polymorphism] is. Beyond that, I’m only dimly able to understand how a given study was conducted and what the authors make of their findings, but not able to speak to the quality of the work. So in this case, I’ll say what a SNP is, because these geneticists can talk of little else, and if you talk genetics, you have to throw in SNP [pronounced "snip"] every so often to qualify yourself. Remember that the genome [the sequence of nucleotides on the chromosomes] fills a pretty large cabinet and makes for some really monotonous reading.
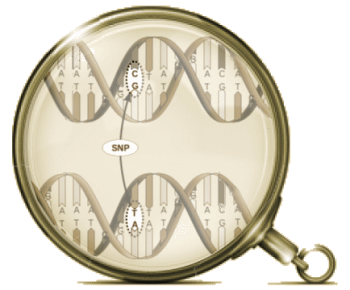
A snip is a place where some segment on the genome differs among individuals or between paired chromosomes by only a single nucleotide. These areas with only one nucleotide difference are easily found and point to areas of variation within and between individuals. Many classic genetic disorders are caused by SNPs [Sickle Cell Anemia, Thalassemia, Cystic Fibrosis, etc]. That’s it for what I know…
The reason for mentioning this study is that this group took what seems to me to be a unique tack in analyzing the data. They identified SNP groupings that were associated with Schizophrenia compared to controls, identifying genotypes [SNP "sets"] that carried a high risk. But then they did something new. They queried a database of Schizophrenic patients who had been in studies where there was a lot of clinical information [eg C.A.T.I.E.]. They classified those cases by phenotype [clinical characteristics].
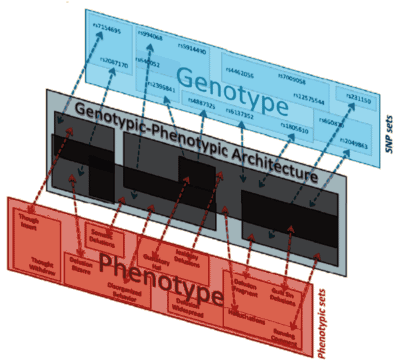
Then they tested the different genotypes [SNP "sets"] against the phenotypes [separated clinically] and found correlations that lead them to conclude that "Schizophrenia is a group of heritable disorders caused by a moderate number of separate genotypic networks associated with several distinct clinical syndromes."
The classification of clinical cases they used was way more complicated that that of the days of Kraepelin or Bleuler, and nothing like the DSM-anything. And their method of correlating genotype [genetic SNP sets] with phenotypes [their clinical classification] was no simple spreadsheet, but rather a complex "architecture" [see their figure above]. It says that Schizophrenia is genetic – that it is not an entity, but many entities [see a guest post from Sandy Steingard…] – "the schizophrenias." The study implies says that the clinical presentation and course of these clinical syndromes define unique "diseases," each with a physical basis. And that’s a mouthful.
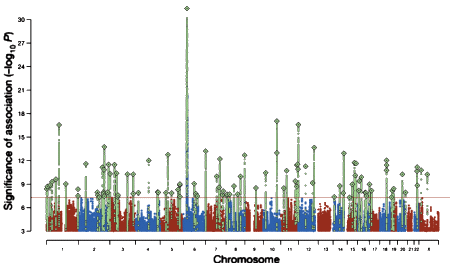
A couple if months ago, there was another Schizophrenia genetic study in Nature that got wide attention [Biological insights from 108 schizophrenia-associated genetic loci] [see Director’s Blog: Mapping the Risk Architecture of Mental Disorders]. The recent write-up of that study in the PsychiatricNews [Dozens of Schizophrenia Risk Loci Identified] was effusive, making analogies to the spires of Dubai and peppered with guest expert speculations. On the other end of the spectrum, Joanna Moncrieff [Critical Psychiatry Network and Mad-in-America] had a different take [A critique of genetic research on schizophrenia – expensive castles in the air], pointing to the massive spending on genetics research with little likely practical yield compared to the lack of attention to the psychosocial needs of these patients. I envy the clarity of such dichotomous convictions. Alas, I always find myself in the middle [in their lifetimes…], and want to insert an AND for every EITHER/OR into these controversies.
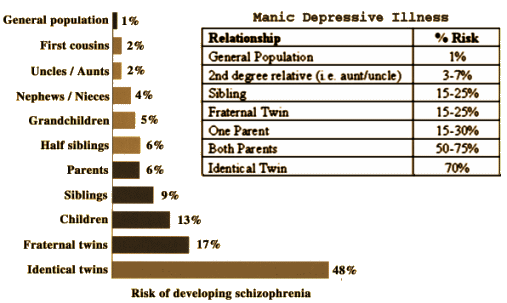
For forty plus years, I’ve seen those slides and figures in books saying that the psychoses have a strong genetic component – and clinical experience has born that out with patients, particularly with Manic-Depressive Illness [classic cases rather than the modern bastardizations]. But just knowing the familial patterns is a factor in diagnosis rather than treatment. For what it’s worth, I believe it, in spite of being of a psychological bent as a clinician. And like almost everyone, when they cracked the genome, I wondered not-if-but-when the genetic associations would get clarified. The early attempts were disappointing. I think the DSM-5 Task Force was counting on such breakthroughs to justify their "biologocal" classification scheme – and the research just didn’t come through, much to their chagrin.
Nurture or Nature?
DNA sequencing became a very big part of my wife’s work in child support early in her career. A blood draw at the time from mom, baby and presumed dad would result in three slides that would be overlapped to show common markers. These essentially were 0’s or 1’s.
Then a variation on our old friend NNT was done to show the probability of two males with the same genetic markers having sex with the mom in the time she was fertile. Excluding identical twins the numbers ran into six figures making it physically impossible for there to be any other possible father.
Great strides have been made in DNA sequencing so that today even minute samples can produce viable results.
My concern is that the KOL’s try for population wide testing and we end up with a Minority Report type of atmosphere where very normal people are categorized as have a mental health issue.
Today there is a push in many parts of government for population wide DNA sampling for such purposes a law enforcement, accident identification, and even determining family relationship.
This could in fact be a big moment in mental health or an opportunity for a profit driven disaster.
Steve Lucas
[… and I cringe in anticipation of the response of the hanger-on-KOL-breakthrough-freaks.] That’s one of your best lines ever, Mickey.
FU to Dr. Carroll, thanks for the hat tip on Snakes in Suits. I got it from Amazon today in a package from Amazon that included Brainless (about “mindless neuroscience”) by Satel and Lillenfeld. Mickey do you know Lillenfeld from your days at Emory? I have a couple of his books.
I strongly recommend the Paul Meehl Reader if you haven’t read it yet. It’s a little drier than the books mentioned above but it is a mile deep with discussions of important topics in psychology/psychiatry still relevant today.
Very cool. Treatment implications?
Treatment implications? Not very close, Tom, as you imply – mainly because the translational steps discussed in the case vignettes were guesswork: educated guesswork but guesswork nonetheless. I don’t fault these investigators for that. If their stuff holds up then it is a definite advance in our understanding. Only the folks Mickey calls the hangers-on KOL crowd and those I call the top-down science bureaucrat crowd like Insel and Hyman are programmed to want to rush to a triumphalist translational victory.
A sobering reminder of the distance between scientific advances and clinical innovation is the case of Huntington disease. The chromosomal locus affected in HD was identified in 1983 and the novel mechanism of trinucleotide repeat polymorphism with pathologic expansions was discovered in 1991. In the interval we have learned a lot about CAG expansions, polyglutamine segments in expressed huntingtin protein, the folding and sheeting of such abnormal proteins, and their interference with cell function. Just knowing the gene, however, has not brought us much closer to understanding the pleomorphic phenotypic (i.e. clinical) expressions of the disease, or the pathophysiologic mechanisms that lead to symptoms, and clinical management has not changed at all, except for genetic testing. The 1983 and 1991 discoveries were epic – it’s just that this is not a simple business. Critics of clinical psychobiology research should keep that in mind just as the KOL crowd of hangers-on and the science bureaucrats should do.
Is KOLCHO the official acronym of KOL crowd of hangers-on?
They remind me of tech and biotech analysts at investment banks. Buy the hype, sell the news….
Just to amplify Dr. Carroll’s point, I went to medical school and did my medicine residency in Memphis Tennessee – a place where Sickle Cell Anemia and the other Hemaglobinopathies are heavily represented. Sickle Cell crises were a common reason to be called to the ER during my years there. Sickle Cell is a genetically transmitted disease where the genetic mechanisms are understood in minute detail, and have been for decades, but the knowledge hasn’t produced a treatment strategy. Sickle Cell Anemia is an autosomal recessive with a single nucleotide polymorphism on chromosome 11 resulting is a single amino acid difference between HgA and HgS. It is known how this difference allows the physical distortion of the molecule that results in a morphologic change in the red cells, and how that results in a physiological change that causes the many clinical problems that come with the affliction. We’ve known all of that throughout my career, but it hasn’t lead to a specific treatment strategy. However, the lifespan and life quality of the patients has been improved substantially by advances in supportive care [not this genetic knowledge].
If this finding in the “schizophrenias” makes it through replication and refinement, who knows where it might lead. But it would be the exception rather than the rule for it to lead us rapidly [if at all] to specific treatment options. However, hope springs eternal…
Dr Mickey
You and your experienced commenters teach. I’m learning, bit by bit, getting the bigger picture, I hope, I think, decades away from the simplistic certainties presented to me as a mom, year after year, by the crowd of KOLs and their hangers-on.
Thankfully yours, Berit
Any research that starts to pick apart the extremely heterogeneous and mystifying label that we call schizophrenia is a good thing. One caveat though: the term genetic disease is extremely loaded. Diseases that result from single SNPs like sickle-cell anemia may be aptly called genetic in their etioloogy (although even this is far from clear cut because symptom severity varies even in these cases).
When it comes to interacting SNPs, the story becomes more complicated by orders of magnitude. Penetrance rates in this study are extremely low, which means that having one of these SNPs sets leads to a higher risk, not a high risk. If the base rate in the general population is 1%, then unless I’m misunderstanding a 70% increase infers a risk of 1.7%. This hardly constitutes a genetic disease, but rather a vulnerability, which is something else altogether. This speaks to the very long road that exists between genotype and phenotype for complex traits, a road that is highly sensitive to environmental context.
So yes, a promising line of enquiry, but not in the simplistic manner suggested by the language used in the paper. I’m not going to hold my breath for a entirely genetic explanation of something that is so clearly moderated by environment.
Adam,
An excellent point! Although it’s an exciting scientific finding [if it holds up], what it means for actual patient care is a great unknown…
Another example would be the initial excitement by some KOLCHOs about cytochromes and SSRIs that came out about ten years ago that seems to have fizzled.
Some genetic research will play out in the end, some will be a rehash of primal scream, recovered memories and other fiascos in psychiatry.
Well, you have to admit NIMH-funded research finally produced something.
Coming soon: Marketing of genetic tests for schizophrenia. Efforts to match each of the 8 genetic types to drugs for treatment. This will produce jimmying of data to promote one drug or another, FDA extensions of patents, and a whole new round of speaking engagements for KOLs! Bonanza!
We’ll all cringe together…
“Coming soon: Marketing of genetic tests for schizophrenia. Efforts to match each of the 8 genetic types to drugs for treatment. This will produce jimmying of data to promote one drug or another, FDA extensions of patents, and a whole new round of speaking engagements for KOLs! Bonanza!”
KOLCHOs have already given their seal of approval:
https://www.youtube.com/watch?v=lsiuxim8vsM
@adam. 70 % oder 1,7 %? From the press release:
“Although individual genes have only weak and inconsistent associations with schizophrenia, groups of interacting gene clusters create an extremely high and consistent risk of illness, on the order of 70 to 100 percent. That makes it almost impossible for people with those genetic variations to avoid the condition. In all, the researchers identified 42 clusters of genetic variations that dramatically increased the risk of schizophrenia.”
http://news.wustl.edu/news/Pages/27358.aspx?utm_source=dlvr.it&utm_medium=tumblr
If this were true, it would be a real sensation.
I think Hans Ulrich got it right.
@Hans
I’ve just had a quick look at the paper and yes, you are certainly right. Thanks for pointing that out. What is claimed is not a 70-100% increase in risk, but an outright 70-100% risk associated with one of these 42 groups. This suggests that the SNP sets are groups of interacting genes that act in concert to create a schizophrenia phenotype in a very large proportion of individuals.
Unfortunately the stats used to calculate this risk are beyond me, and there’s no clear articulation in the paper of how that risk was calculated. It seems that they compared the frequency of the SNP sets in both clinical and control groups to determine risk. Does anyone know how they did that with such a small control group?
Here’s my beef: There is an extremely low statistical likelihood of finding any of these SNP sets in control group (<1%) and also a reasonably small number of participants with any single SNP set in the clinical group (20-200 or .5-5%). Taken together, this suggests that any SNP set would have an extremely low frequency in the control group (between .2 and 2 participants). Given that they have a fairly small control group, I would like to see how they calculated these risks. Any stats wizards out there?
All that aside, the association with patterns of clinical symptoms they report is really quite AMAZING, and I think that is by far the most significant part of the research. In terms of risk factors, I don't mean to come across as a killjoy, but I've been conditioned by so many worthless and unreplicable GWAS studies to always be a bit sceptical.
I forgot to mention this: there was only a single SNP set that had 100% risk, and it was present in only 9/3800 participants…..
So amazing I’m wondering exactly what subjective criteria were used to classify those symptoms….
Jeffrey A. Lieberman, MD, chair of the Department of Psychiatry at Columbia University College of Physicians and Surgeons and Psychiatrist-in-Chief at New York Presbyterian Hospital–Columbia University Medical Center, New York City, told Medscape Medical News:
“I think it is interesting to suggest that there may be symptomatic phenotypes that correlate with a specific pattern of genetic anomalies, but I would want to see some replication before I would be willing to accept the finding.”
http://www.medscape.com/viewarticle/832018
Now that he’s an ex-President he seems to be out of millions of saved lives hype mode and into clinical restraint mode.
In a related matter of good clinical judgment, I want some more proof that giving SDAs to nonpsychotic people is actually “neuroprotective” and not neurotoxic.
At least the blood test isn’t hurting anyone except financially.